ProSight Native
Ultrafast intact mass profiling and targeted top-down mass spectrometry analysis.
Take your intact mass analysis to the next level.
Our easy-to-use interface makes deconvoluting spectra simple. Plus, our ultra-fast algorithms ensure you won't have to sit around and wait for results. ProSight Native has simple user interfaces for interacting with data. Our deconvolution is extremely fast and can process protein data across a wide range of molecular weights, from peptides to protein therapeutics to AAVs.
Interacting with your data is easy. Simply select the portion of the chromatogram to average and the deconvolution results will instantly show up. Here, the deconvolution results of Orbitrap data containing multiple ribosomal proteins is shown.
Simplify complexity.
Unveil the full composition and stoichiometry of a protein complex using ProSight Native’s unique workflow to analyze each level of a complex-down mass spectrometry experiment:
Intact complex mass analysis
Subunit mass analysis
Subunit fragmentation analysis.
ProSight Native is currently the only software available that can perform all 3 analysis components.
Features
Determine.
Assess protein complex and subunit masses with mass deconvolution algorithms. ProSight Native integrates our flagship ProSight database search engine, allowing you to then elucidate subunit proteoform identities.
Automatically compile data to yield the full stoichiometry of subunits and even non-covalently-bound ligands associated with the complex.
Deconvolute.
Determine masses for isotopically-resolved and -unresolved proteins. ProSight Native combines kDecon and THRASH deconvolution for analyzing proteins of any size.
Search.
Search native proteoform fragmentation data against candidate proteoforms using ProSight search. Candidates are composed of evidence-backed, site-specific post-translational modifications from UniProt XML files.
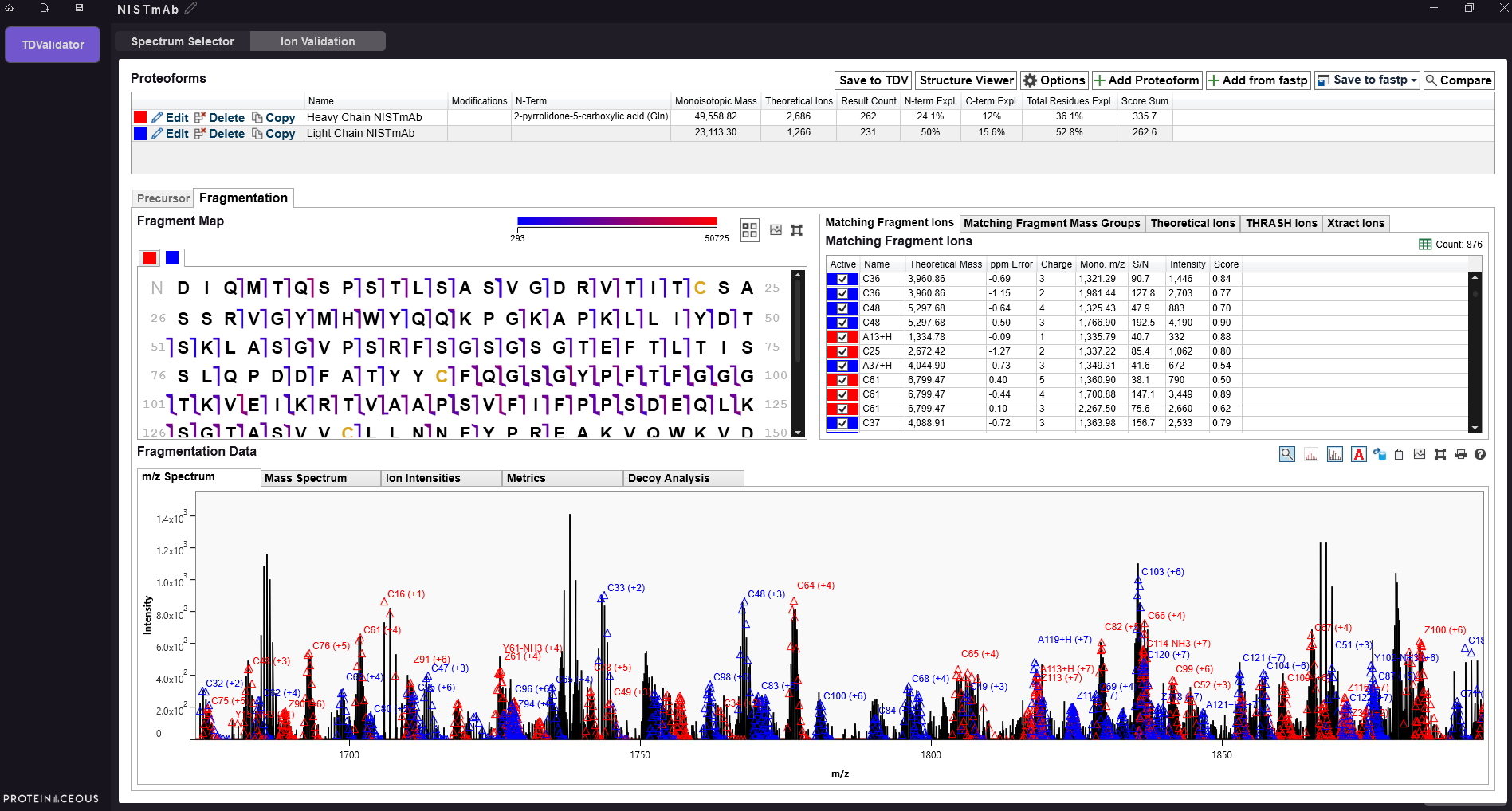
Simplify Top-Down Analysis with TDValidator.
Straightforward proteoform characterization is now readily accessible. We have integrated TDValidator into ProSight Native and added a host of new features, including a simplified file browser for selecting and averaging fragmentation spectra.
Antibody sequencing is performed in the TDValidator module of ProSight Native. Thorough coverage of both the heavy and light chain of an antibody was achieved.
Analyze.
Perform targeted analysis of a single proteoform or multiple proteoforms.
Input proteoforms for a single fragment spectrum and match all of the theoretical fragment ions to the spectral data.
Match precursor ions to precursor spectral data.
Perform fragmentation matching on data from a wide range of fragmentation techniques, including CID, HCD, ETD, ECD, EThcD, ETciD, and UVPD.
Visualize.
Characterize proteoforms using fragmentation evidence.
Analyze fragmentation spectra and corresponding matching ions to confidently localize proteoform modifications.
Overlay matching ions from different proteoforms on the same spectra and uses a host of visualization tools and metrics to power in-depth study of proteoforms.
Look for internal fragment ions formed during fragmentation processes. The ions can be visualized by color gradient heat map on the fragment map itself. Internal fragment ions can provide additional evidence for modification site localization, increasing confidence in assignments.
Validate.
Assess fragmentation coverage using fragmentation maps and related metrics.
Validate fragmentation ion matches to confirm post-translational modification site assignments.
Get ProSight Native
To receive a free 60-day demo license or purchase a subscription, please contact us. In your request, please indicate your title and institution.